Note
Go to the end to download the full example code.
Gibbs sampling#
This tutorial shows how to use CUQIpy to perform Gibbs sampling. Gibbs sampling is a Markov chain Monte Carlo (MCMC) method for sampling a joint probability distribution.
Opposed to jointly sampling the distribution simultaneously, Gibbs sampling samples the variables of the distribution sequentially, one variable at a time. When a variable represents a random vector, the whole vector is sampled simultaneously.
The sampling of each variable is done by sampling from the conditional distribution of that variable given (fixed, previously sampled) values of the other variables.
This is often a very efficient way of sampling from a joint distribution if the conditional distributions are easy to sample from. This is one way to exploit the structure of the joint distribution. On the other hand, if the conditional distributions are highly correlated and/or are difficult to sample from, then Gibbs sampling can be very inefficient.
For these reasons, Gibbs sampling is often a double-edged sword, that needs to be used in the right context.
Setup#
We start by importing the necessary modules
import numpy as np
import matplotlib.pyplot as plt
from cuqi.testproblem import Deconvolution1D
from cuqi.distribution import Gaussian, Gamma, JointDistribution, GMRF, LMRF
from cuqi.legacy.sampler import Gibbs, LinearRTO, Conjugate, UGLA, ConjugateApprox
np.random.seed(0)
Forward model and data#
We define the forward model and data. Here we use a 1D deconvolution problem, so the forward model is linear, that is:
where \(\mathbf{A}\) is the convolution matrix, and \(\mathbf{x}\) is the input signal.
We load this example from the testproblem library of CUQIpy and visualize the true solution (sharp signal) and data (convolved signal).
# Model and data
A, y_obs, probinfo = Deconvolution1D(phantom='square').get_components()
# Get dimension of signal
n = A.domain_dim
# Plot exact solution and observed data
plt.subplot(121)
probinfo.exactSolution.plot()
plt.title('exact solution')
plt.subplot(122)
y_obs.plot()
plt.title("Observed data")
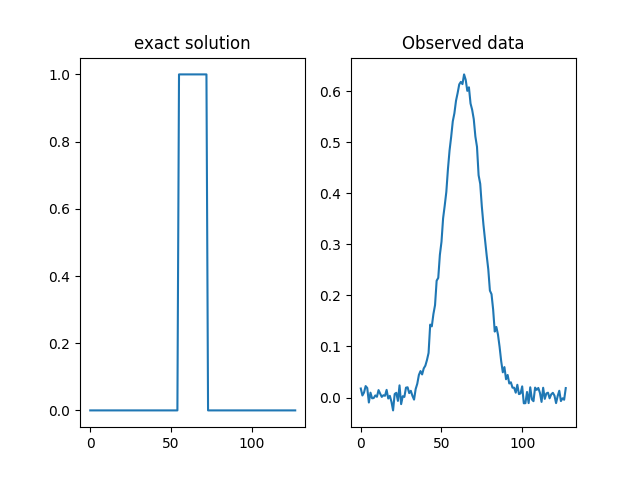
Text(0.5, 1.0, 'Observed data')
Hierarchical Bayesian model#
We define the following hierarchical model:
where \(\mathbf{y}\) is the observed data, and \(\mathbf{x}\) is the unknown signal. The hyperparameters \(d\) and \(l\) are the precision of the prior distribution of \(\mathbf{x}\) and the noise, respectively.
The prior distribution of \(\mathbf{x}\) is a Gaussian Markov random field (GMRF) with zero mean and precision \(d\). It can be viewed as a Gaussian prior on the differences between neighboring elements of \(\mathbf{x}\).
In CUQIpy the model can be defined as follows:
# Define distributions
d = Gamma(1, 1e-4)
l = Gamma(1, 1e-4)
x = GMRF(np.zeros(n), lambda d: d)
y = Gaussian(A, lambda l: 1/l)
# Combine into a joint distribution
joint = JointDistribution(d, l, x, y)
# View the joint distribution
print(joint)
JointDistribution(
Equation:
p(d,l,x,y) = p(d)p(l)p(x|d)p(y|x,l)
Densities:
d ~ CUQI Gamma.
l ~ CUQI Gamma.
x ~ CUQI GMRF. Conditioning variables ['d'].
y ~ CUQI Gaussian. Conditioning variables ['x', 'l'].
)
Notice that the joint distribution prints a mathematical expression for the density functions that make up \(p(d,l,\mathbf{x},\mathbf{y})\). In this case they are all distributions, but this need not be the case.
Defining the posterior distribution#
Now we define the posterior distribution, which is the joint distribution conditioned on the observed data. That is, \(p(d, l, \mathbf{x} \mid \mathbf{y}=\mathbf{y}_\mathrm{obs})\)
This is done in the following way:
# Define posterior by conditioning on the data
posterior = joint(y=y_obs)
# View the structure of the posterior
print(posterior)
JointDistribution(
Equation:
p(d,l,x|y) ∝ p(d)p(l)p(x|d)L(x,l|y)
Densities:
d ~ CUQI Gamma.
l ~ CUQI Gamma.
x ~ CUQI GMRF. Conditioning variables ['d'].
y ~ CUQI Gaussian Likelihood function. Parameters ['x', 'l'].
)
Notice that after conditioning on the data, the distribution associated with \(\mathbf{y}\) became a likelihood function and that the posterior is now a joint distribution of the variables \(d\), \(l\), \(\mathbf{x}\).
Gibbs Sampler#
The hierarchical model above has some important properties that we can exploit to make the sampling more efficient. First, note that the Gamma distribution are conjugate priors for the precision of the Gaussian distributions. This means that we can efficiently sample from \(d\) and \(l\) conditional on the other variables.
Second, note that the prior distribution of \(\mathbf{x}\) is
a Gaussian Markov random field (GMRF) and that the distribution for
\(\mathbf{y}\) is also Gaussian with a Linear operator acting
on \(\mathbf{x}\) as the mean variable. This means that we can
efficiently sample from \(\mathbf{x}\) conditional on the other
variables using the LinearRTO sampler.
Taking these two facts into account, we can define a Gibbs sampler
that uses the Conjugate sampler for \(d\) and \(l\) and
the LinearRTO sampler for \(\mathbf{x}\).
This is done in CUQIpy as follows:
# Define sampling strategy
sampling_strategy = {
'x': LinearRTO,
'd': Conjugate,
'l': Conjugate
}
# Define Gibbs sampler
sampler = Gibbs(posterior, sampling_strategy)
# Run sampler
samples = sampler.sample(Ns=1000, Nb=200)
/home/runner/work/CUQIpy/CUQIpy/demos/tutorials/Gibbs.py:167: UserWarning:
You are using the legacy sampler 'Gibbs'.
This will be removed in a future release of CUQIpy.
Please consider using the new samplers in the 'cuqi.sampler' module.
sampler = Gibbs(posterior, sampling_strategy)
/home/runner/work/CUQIpy/CUQIpy/cuqi/legacy/sampler/_rto.py:77: UserWarning:
You are using the legacy sampler 'LinearRTO'.
This will be removed in a future release of CUQIpy.
Please consider using the new samplers in the 'cuqi.sampler' module.
super().__init__(target, x0=x0, **kwargs)
Warmup 2 / 200
Warmup 4 / 200
Warmup 6 / 200
Warmup 8 / 200
Warmup 10 / 200
Warmup 12 / 200
Warmup 14 / 200
Warmup 16 / 200
Warmup 18 / 200
Warmup 20 / 200
Warmup 22 / 200
Warmup 24 / 200
Warmup 26 / 200
Warmup 28 / 200
Warmup 30 / 200
Warmup 32 / 200
Warmup 34 / 200
Warmup 36 / 200
Warmup 38 / 200
Warmup 40 / 200
Warmup 42 / 200
Warmup 44 / 200
Warmup 46 / 200
Warmup 48 / 200
Warmup 50 / 200
Warmup 52 / 200
Warmup 54 / 200
Warmup 56 / 200
Warmup 58 / 200
Warmup 60 / 200
Warmup 62 / 200
Warmup 64 / 200
Warmup 66 / 200
Warmup 68 / 200
Warmup 70 / 200
Warmup 72 / 200
Warmup 74 / 200
Warmup 76 / 200
Warmup 78 / 200
Warmup 80 / 200
Warmup 82 / 200
Warmup 84 / 200
Warmup 86 / 200
Warmup 88 / 200
Warmup 90 / 200
Warmup 92 / 200
Warmup 94 / 200
Warmup 96 / 200
Warmup 98 / 200
Warmup 100 / 200
Warmup 102 / 200
Warmup 104 / 200
Warmup 106 / 200
Warmup 108 / 200
Warmup 110 / 200
Warmup 112 / 200
Warmup 114 / 200
Warmup 116 / 200
Warmup 118 / 200
Warmup 120 / 200
Warmup 122 / 200
Warmup 124 / 200
Warmup 126 / 200
Warmup 128 / 200
Warmup 130 / 200
Warmup 132 / 200
Warmup 134 / 200
Warmup 136 / 200
Warmup 138 / 200
Warmup 140 / 200
Warmup 142 / 200
Warmup 144 / 200
Warmup 146 / 200
Warmup 148 / 200
Warmup 150 / 200
Warmup 152 / 200
Warmup 154 / 200
Warmup 156 / 200
Warmup 158 / 200
Warmup 160 / 200
Warmup 162 / 200
Warmup 164 / 200
Warmup 166 / 200
Warmup 168 / 200
Warmup 170 / 200
Warmup 172 / 200
Warmup 174 / 200
Warmup 176 / 200
Warmup 178 / 200
Warmup 180 / 200
Warmup 182 / 200
Warmup 184 / 200
Warmup 186 / 200
Warmup 188 / 200
Warmup 190 / 200
Warmup 192 / 200
Warmup 194 / 200
Warmup 196 / 200
Warmup 198 / 200
Warmup 200 / 200
Warmup 200 / 200
Sample 10 / 1000
Sample 20 / 1000
Sample 30 / 1000
Sample 40 / 1000
Sample 50 / 1000
Sample 60 / 1000
Sample 70 / 1000
Sample 80 / 1000
Sample 90 / 1000
Sample 100 / 1000
Sample 110 / 1000
Sample 120 / 1000
Sample 130 / 1000
Sample 140 / 1000
Sample 150 / 1000
Sample 160 / 1000
Sample 170 / 1000
Sample 180 / 1000
Sample 190 / 1000
Sample 200 / 1000
Sample 210 / 1000
Sample 220 / 1000
Sample 230 / 1000
Sample 240 / 1000
Sample 250 / 1000
Sample 260 / 1000
Sample 270 / 1000
Sample 280 / 1000
Sample 290 / 1000
Sample 300 / 1000
Sample 310 / 1000
Sample 320 / 1000
Sample 330 / 1000
Sample 340 / 1000
Sample 350 / 1000
Sample 360 / 1000
Sample 370 / 1000
Sample 380 / 1000
Sample 390 / 1000
Sample 400 / 1000
Sample 410 / 1000
Sample 420 / 1000
Sample 430 / 1000
Sample 440 / 1000
Sample 450 / 1000
Sample 460 / 1000
Sample 470 / 1000
Sample 480 / 1000
Sample 490 / 1000
Sample 500 / 1000
Sample 510 / 1000
Sample 520 / 1000
Sample 530 / 1000
Sample 540 / 1000
Sample 550 / 1000
Sample 560 / 1000
Sample 570 / 1000
Sample 580 / 1000
Sample 590 / 1000
Sample 600 / 1000
Sample 610 / 1000
Sample 620 / 1000
Sample 630 / 1000
Sample 640 / 1000
Sample 650 / 1000
Sample 660 / 1000
Sample 670 / 1000
Sample 680 / 1000
Sample 690 / 1000
Sample 700 / 1000
Sample 710 / 1000
Sample 720 / 1000
Sample 730 / 1000
Sample 740 / 1000
Sample 750 / 1000
Sample 760 / 1000
Sample 770 / 1000
Sample 780 / 1000
Sample 790 / 1000
Sample 800 / 1000
Sample 810 / 1000
Sample 820 / 1000
Sample 830 / 1000
Sample 840 / 1000
Sample 850 / 1000
Sample 860 / 1000
Sample 870 / 1000
Sample 880 / 1000
Sample 890 / 1000
Sample 900 / 1000
Sample 910 / 1000
Sample 920 / 1000
Sample 930 / 1000
Sample 940 / 1000
Sample 950 / 1000
Sample 960 / 1000
Sample 970 / 1000
Sample 980 / 1000
Sample 990 / 1000
Sample 1000 / 1000
Sample 1000 / 1000
Analyze results#
After sampling we can inspect the results. The samples are stored as a dictionary with the variable names as keys. Samples for each variable is stored as a CUQIpy Samples object which contains the many convenience methods for diagnostics and plotting of MCMC samples.
# Plot credible intervals for the signal
samples['x'].plot_ci(exact=probinfo.exactSolution)
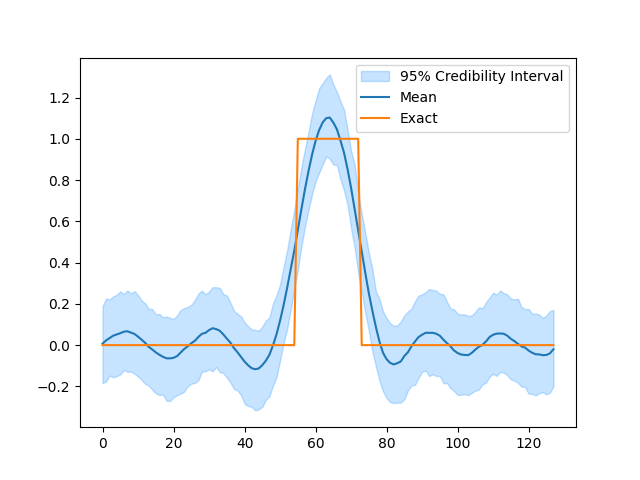
[<matplotlib.lines.Line2D object at 0x7f7b035276e0>, <matplotlib.lines.Line2D object at 0x7f7b0349aa80>, <matplotlib.collections.FillBetweenPolyCollection object at 0x7f7b036336e0>]
Trace plot for d
samples['d'].plot_trace(figsize=(8,2))
array([[<Axes: title={'center': 'd'}>, <Axes: title={'center': 'd'}>]],
dtype=object)
Trace plot for l
samples['l'].plot_trace(figsize=(8,2))
array([[<Axes: title={'center': 'l'}>, <Axes: title={'center': 'l'}>]],
dtype=object)
Switching to a piecewise constant prior#
Notice that while the sampling went well in the previous example, the posterior distribution did not match the characteristics of the exact solution. We can improve this result by switching to a prior that better matches the exact solution \(\mathbf{x}\).
One choice is the Laplace difference prior, which assumes a Laplace distribution for the differences between neighboring elements of \(\mathbf{x}\). That is,
which means that \(x_i-x_{i-1} \sim \mathrm{Laplace}(0, d^{-1})\).
This prior is implemented in CUQIpy as the LMRF distribution.
To update our model we simply need to replace the GMRF distribution
with the LMRF distribution. Note that the Laplace distribution
is defined via a scale parameter, so we invert the parameter \(d\).
This laplace distribution and new posterior can be defined as follows:
# Define new distribution for x
x = LMRF(0, lambda d: 1/d, geometry=n)
# Define new joint distribution with piecewise constant prior
joint_Ld = JointDistribution(d, l, x, y)
# Define new posterior by conditioning on the data
posterior_Ld = joint_Ld(y=y_obs)
print(posterior_Ld)
JointDistribution(
Equation:
p(d,l,x|y) ∝ p(d)p(l)p(x|d)L(x,l|y)
Densities:
d ~ CUQI Gamma.
l ~ CUQI Gamma.
x ~ CUQI LMRF. Conditioning variables ['d'].
y ~ CUQI Gaussian Likelihood function. Parameters ['x', 'l'].
)
Gibbs Sampler (with Laplace prior)#
Using the same approach as earlier we can define a Gibbs sampler
for this new hierarchical model. The only difference is that we
now need to use a different sampler for \(\mathbf{x}\) because
the LinearRTO sampler only works for Gaussian distributions.
In this case we use the UGLA (Unadjusted Gaussian Laplace Approximation) sampler for \(\mathbf{x}\). We also use an approximate Conjugate sampler for \(d\) which approximately samples from the posterior distribution of \(d\) conditional on the other variables in an efficient manner. For more details see e.g. this paper <https://arxiv.org/abs/2104.06919>.
# Define sampling strategy
sampling_strategy = {
'x': UGLA,
'd': ConjugateApprox,
'l': Conjugate
}
# Define Gibbs sampler
sampler_Ld = Gibbs(posterior_Ld, sampling_strategy)
# Run sampler
samples_Ld = sampler_Ld.sample(Ns=1000, Nb=200)
/home/runner/work/CUQIpy/CUQIpy/demos/tutorials/Gibbs.py:254: UserWarning:
You are using the legacy sampler 'Gibbs'.
This will be removed in a future release of CUQIpy.
Please consider using the new samplers in the 'cuqi.sampler' module.
sampler_Ld = Gibbs(posterior_Ld, sampling_strategy)
/home/runner/work/CUQIpy/CUQIpy/cuqi/legacy/sampler/_laplace_approximation.py:57: UserWarning:
You are using the legacy sampler 'UGLA'.
This will be removed in a future release of CUQIpy.
Please consider using the new samplers in the 'cuqi.sampler' module.
super().__init__(target, x0=x0, **kwargs)
Warmup 2 / 200
Warmup 4 / 200
Warmup 6 / 200
Warmup 8 / 200
Warmup 10 / 200
Warmup 12 / 200
Warmup 14 / 200
Warmup 16 / 200
Warmup 18 / 200
Warmup 20 / 200
Warmup 22 / 200
Warmup 24 / 200
Warmup 26 / 200
Warmup 28 / 200
Warmup 30 / 200
Warmup 32 / 200
Warmup 34 / 200
Warmup 36 / 200
Warmup 38 / 200
Warmup 40 / 200
Warmup 42 / 200
Warmup 44 / 200
Warmup 46 / 200
Warmup 48 / 200
Warmup 50 / 200
Warmup 52 / 200
Warmup 54 / 200
Warmup 56 / 200
Warmup 58 / 200
Warmup 60 / 200
Warmup 62 / 200
Warmup 64 / 200
Warmup 66 / 200
Warmup 68 / 200
Warmup 70 / 200
Warmup 72 / 200
Warmup 74 / 200
Warmup 76 / 200
Warmup 78 / 200
Warmup 80 / 200
Warmup 82 / 200
Warmup 84 / 200
Warmup 86 / 200
Warmup 88 / 200
Warmup 90 / 200
Warmup 92 / 200
Warmup 94 / 200
Warmup 96 / 200
Warmup 98 / 200
Warmup 100 / 200
Warmup 102 / 200
Warmup 104 / 200
Warmup 106 / 200
Warmup 108 / 200
Warmup 110 / 200
Warmup 112 / 200
Warmup 114 / 200
Warmup 116 / 200
Warmup 118 / 200
Warmup 120 / 200
Warmup 122 / 200
Warmup 124 / 200
Warmup 126 / 200
Warmup 128 / 200
Warmup 130 / 200
Warmup 132 / 200
Warmup 134 / 200
Warmup 136 / 200
Warmup 138 / 200
Warmup 140 / 200
Warmup 142 / 200
Warmup 144 / 200
Warmup 146 / 200
Warmup 148 / 200
Warmup 150 / 200
Warmup 152 / 200
Warmup 154 / 200
Warmup 156 / 200
Warmup 158 / 200
Warmup 160 / 200
Warmup 162 / 200
Warmup 164 / 200
Warmup 166 / 200
Warmup 168 / 200
Warmup 170 / 200
Warmup 172 / 200
Warmup 174 / 200
Warmup 176 / 200
Warmup 178 / 200
Warmup 180 / 200
Warmup 182 / 200
Warmup 184 / 200
Warmup 186 / 200
Warmup 188 / 200
Warmup 190 / 200
Warmup 192 / 200
Warmup 194 / 200
Warmup 196 / 200
Warmup 198 / 200
Warmup 200 / 200
Warmup 200 / 200
Sample 10 / 1000
Sample 20 / 1000
Sample 30 / 1000
Sample 40 / 1000
Sample 50 / 1000
Sample 60 / 1000
Sample 70 / 1000
Sample 80 / 1000
Sample 90 / 1000
Sample 100 / 1000
Sample 110 / 1000
Sample 120 / 1000
Sample 130 / 1000
Sample 140 / 1000
Sample 150 / 1000
Sample 160 / 1000
Sample 170 / 1000
Sample 180 / 1000
Sample 190 / 1000
Sample 200 / 1000
Sample 210 / 1000
Sample 220 / 1000
Sample 230 / 1000
Sample 240 / 1000
Sample 250 / 1000
Sample 260 / 1000
Sample 270 / 1000
Sample 280 / 1000
Sample 290 / 1000
Sample 300 / 1000
Sample 310 / 1000
Sample 320 / 1000
Sample 330 / 1000
Sample 340 / 1000
Sample 350 / 1000
Sample 360 / 1000
Sample 370 / 1000
Sample 380 / 1000
Sample 390 / 1000
Sample 400 / 1000
Sample 410 / 1000
Sample 420 / 1000
Sample 430 / 1000
Sample 440 / 1000
Sample 450 / 1000
Sample 460 / 1000
Sample 470 / 1000
Sample 480 / 1000
Sample 490 / 1000
Sample 500 / 1000
Sample 510 / 1000
Sample 520 / 1000
Sample 530 / 1000
Sample 540 / 1000
Sample 550 / 1000
Sample 560 / 1000
Sample 570 / 1000
Sample 580 / 1000
Sample 590 / 1000
Sample 600 / 1000
Sample 610 / 1000
Sample 620 / 1000
Sample 630 / 1000
Sample 640 / 1000
Sample 650 / 1000
Sample 660 / 1000
Sample 670 / 1000
Sample 680 / 1000
Sample 690 / 1000
Sample 700 / 1000
Sample 710 / 1000
Sample 720 / 1000
Sample 730 / 1000
Sample 740 / 1000
Sample 750 / 1000
Sample 760 / 1000
Sample 770 / 1000
Sample 780 / 1000
Sample 790 / 1000
Sample 800 / 1000
Sample 810 / 1000
Sample 820 / 1000
Sample 830 / 1000
Sample 840 / 1000
Sample 850 / 1000
Sample 860 / 1000
Sample 870 / 1000
Sample 880 / 1000
Sample 890 / 1000
Sample 900 / 1000
Sample 910 / 1000
Sample 920 / 1000
Sample 930 / 1000
Sample 940 / 1000
Sample 950 / 1000
Sample 960 / 1000
Sample 970 / 1000
Sample 980 / 1000
Sample 990 / 1000
Sample 1000 / 1000
Sample 1000 / 1000
Analyze results#
Again we can inspect the results. Here we notice the posterior distribution matches the exact solution much better.
# Plot credible intervals for the signal
samples_Ld['x'].plot_ci(exact=probinfo.exactSolution)
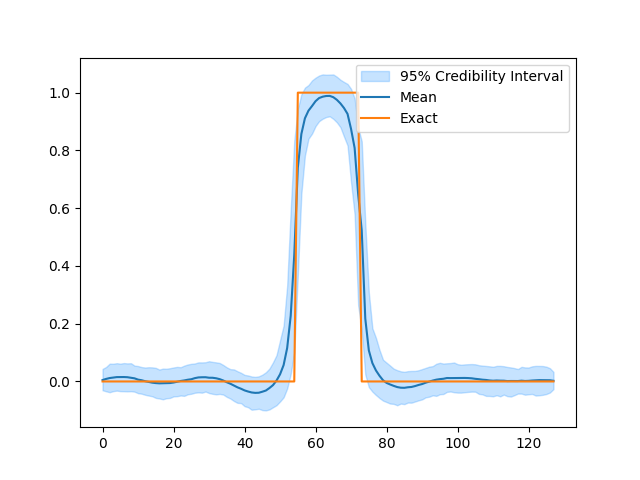
[<matplotlib.lines.Line2D object at 0x7f7b011a9550>, <matplotlib.lines.Line2D object at 0x7f7b011a97c0>, <matplotlib.collections.FillBetweenPolyCollection object at 0x7f7b034c1160>]
samples_Ld['d'].plot_trace(figsize=(8,2))
array([[<Axes: title={'center': 'd'}>, <Axes: title={'center': 'd'}>]],
dtype=object)
samples_Ld['l'].plot_trace(figsize=(8,2))
array([[<Axes: title={'center': 'l'}>, <Axes: title={'center': 'l'}>]],
dtype=object)
Total running time of the script: (0 minutes 32.014 seconds)
